Introduction to Digital PCR
Digital PCR (dPCR) is the third-generation PCR technology developed after conventional PCR and real-time quantitative PCR (qPCR). Its core principle is to partition a reaction mixture containing target nucleic acid molecules into tens of thousands of independent micro-reaction units (such as micro-droplets or microwells), with individual PCR amplification occurring within each unit.
After amplification is complete, an endpoint detection (typically of a fluorescence signal) is performed on each unit. The presence or absence of a signal determines whether that unit contains the target nucleic acid molecule. By counting the number of positive units and applying Poisson distribution statistics, the absolute copy number of the target nucleic acid in the original sample can be directly calculated.
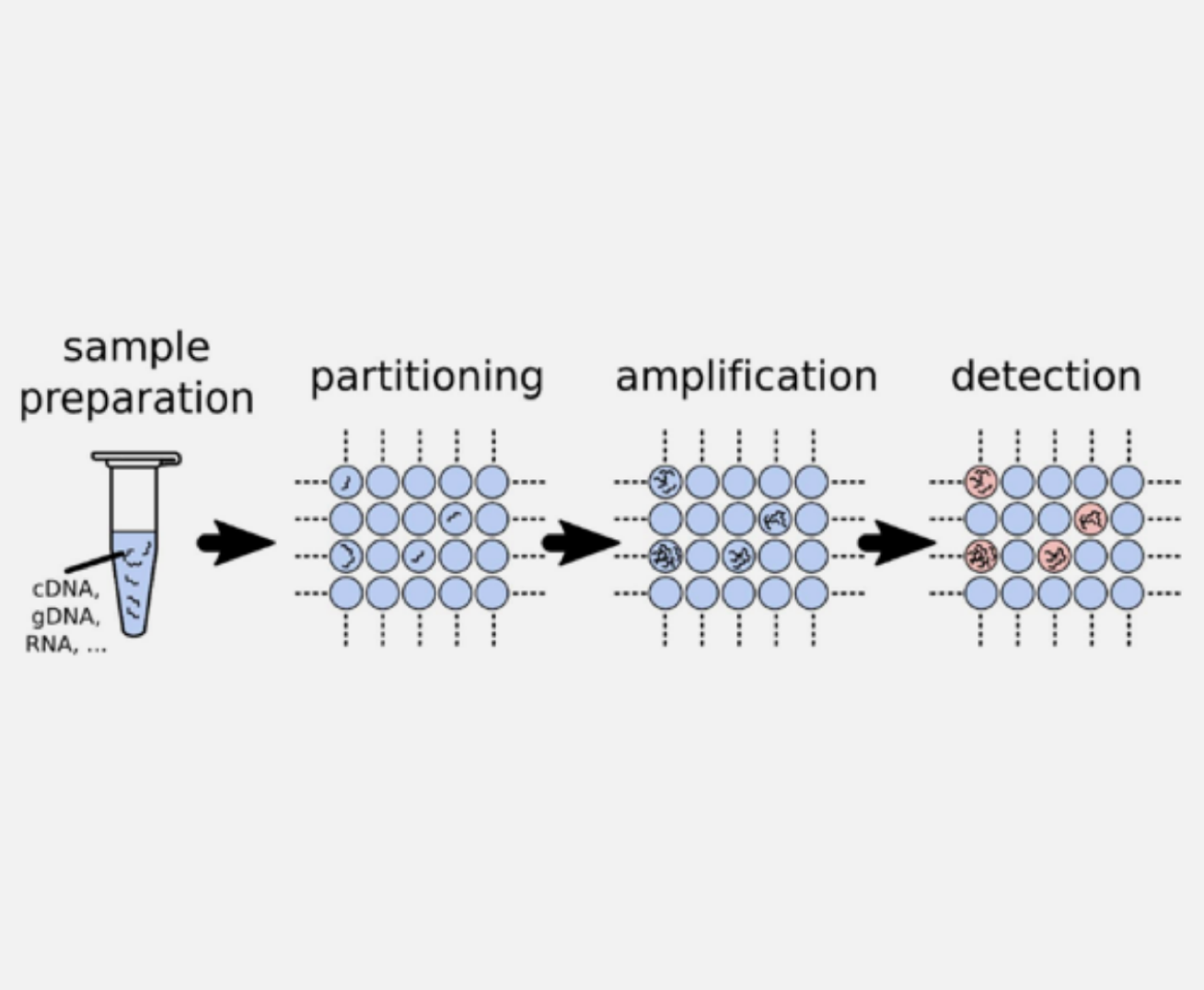
The core advantage of Digital PCR lies in its capability for absolute quantification without reliance on a standard curve, leading to more precise and reliable results. Additionally, it has a higher tolerance to inhibitors and demonstrates excellent sensitivity in detecting targets of extremely low abundance, such as rare mutations, trace pathogens, and circulating tumor DNA (ctDNA).
These features have enabled dPCR to show tremendous application value and potential in life science research and clinical diagnostics, including fields such as tumor liquid biopsy, non-invasive prenatal diagnosis, precise pathogen detection, gene expression analysis, quantification of transgenic components, and the valuation of reference standards.
Types of Digital PCR
Digital PCR can be broadly classified into three main types based on the partitioning method used to separate the sample into many individual reaction units:
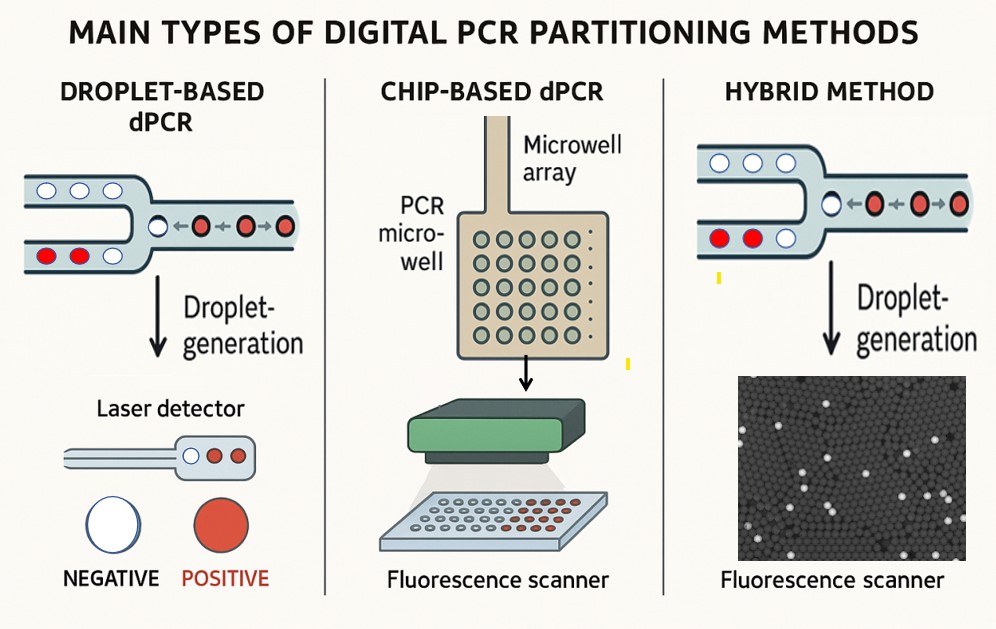
Droplet-based digital PCR
This method partitions the PCR mixture into tens of thousands tiny water-in-oil droplets, each acting as an independent PCR microreactor. Droplets are generated using microfluidics and then subjected to PCR amplification. After amplification, droplets are analyzed individually for fluorescence to determine positive or negative partitions. This approach offers a very high number of partitions and is widely used for its sensitivity and precision.
Chip-based digital PCR
Hybrid methods
Each partitioning strategy isolates nucleic acid molecules into separate reaction compartments, enabling the use of Poisson statistics to accurately quantify target DNA or RNA molecules by counting positive and negative partitions. The choice of partitioning method affects the number of partitions, partition volume, workflow complexity, and instrument requirements.
Poisson statistics in digital PCR
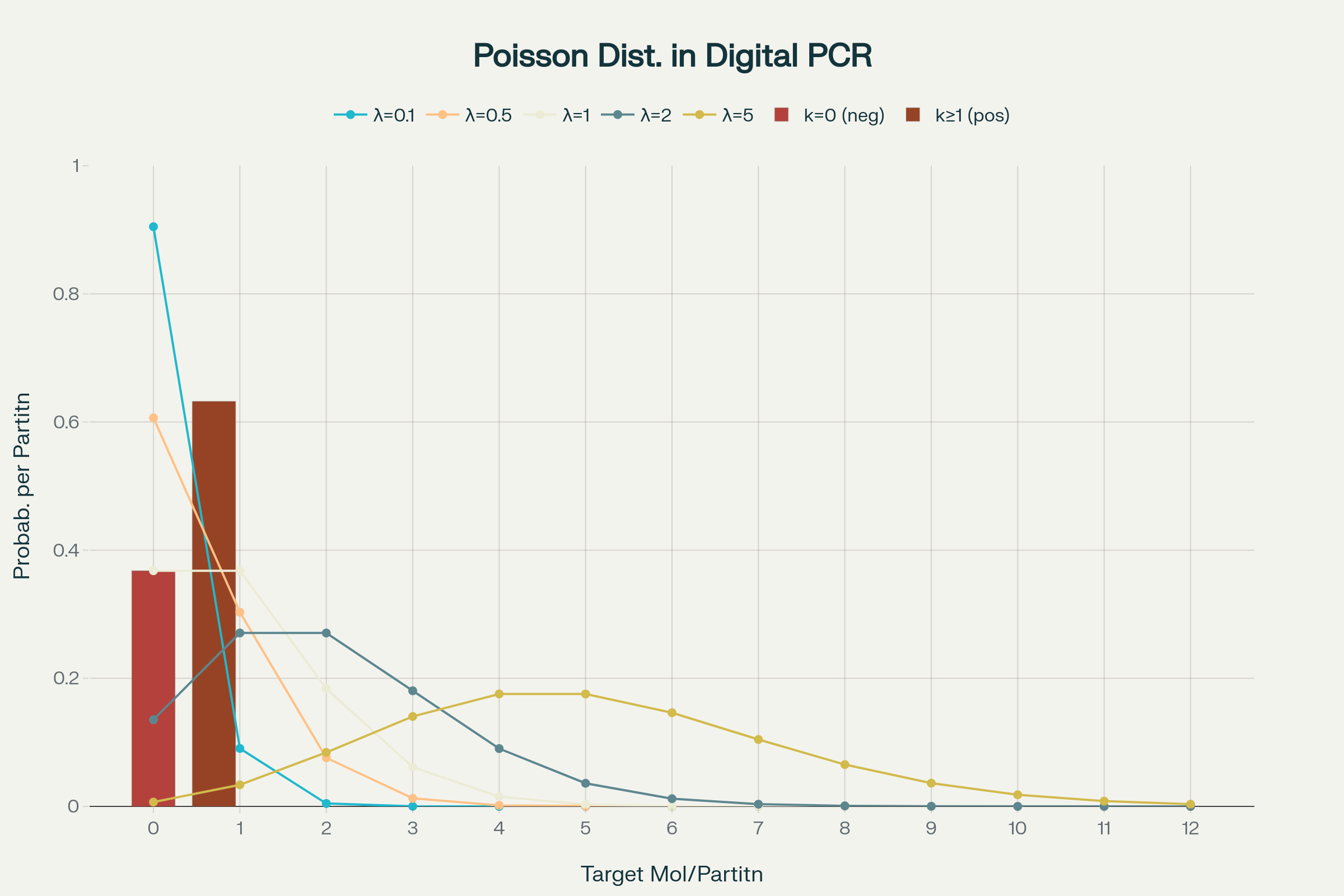
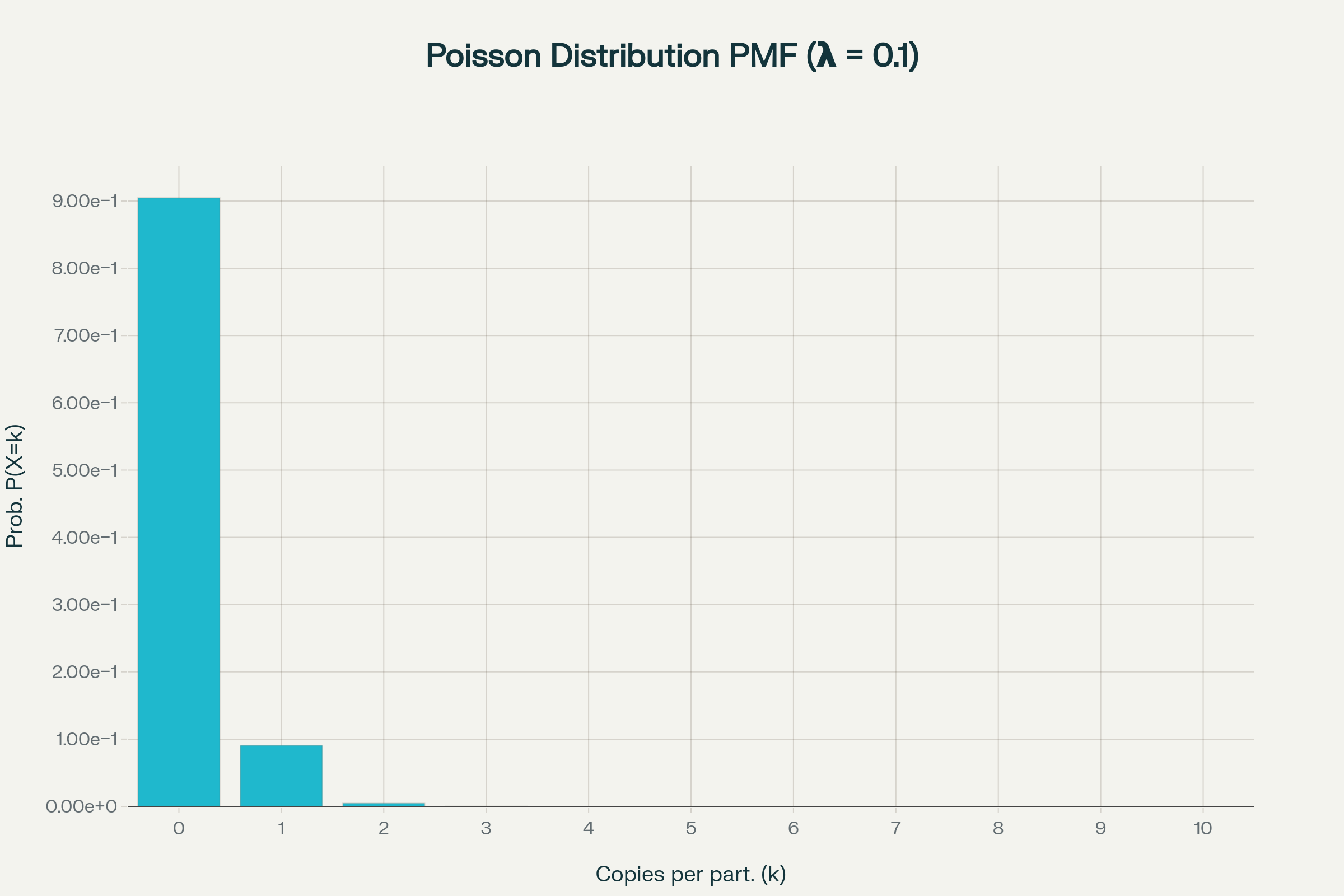
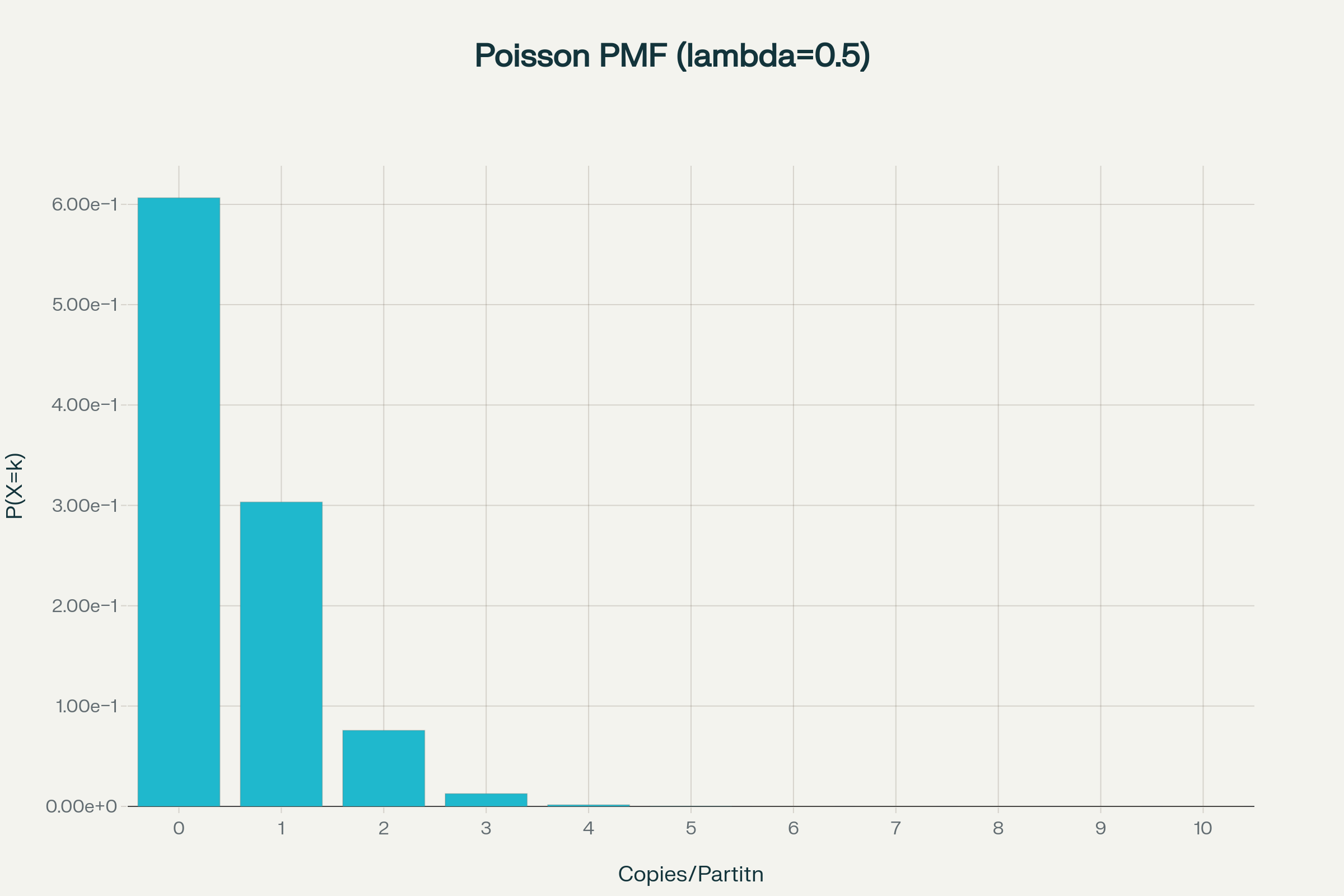
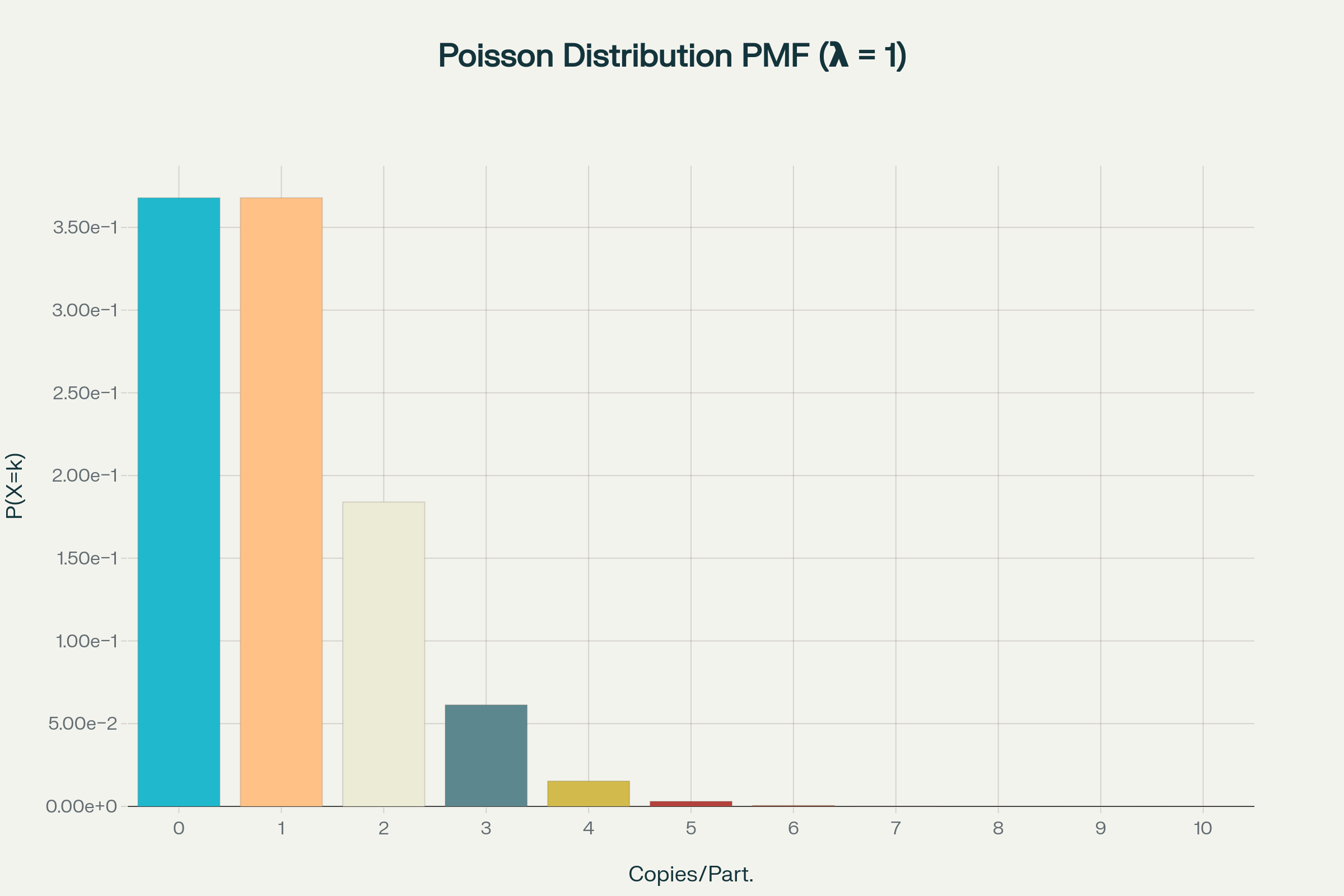
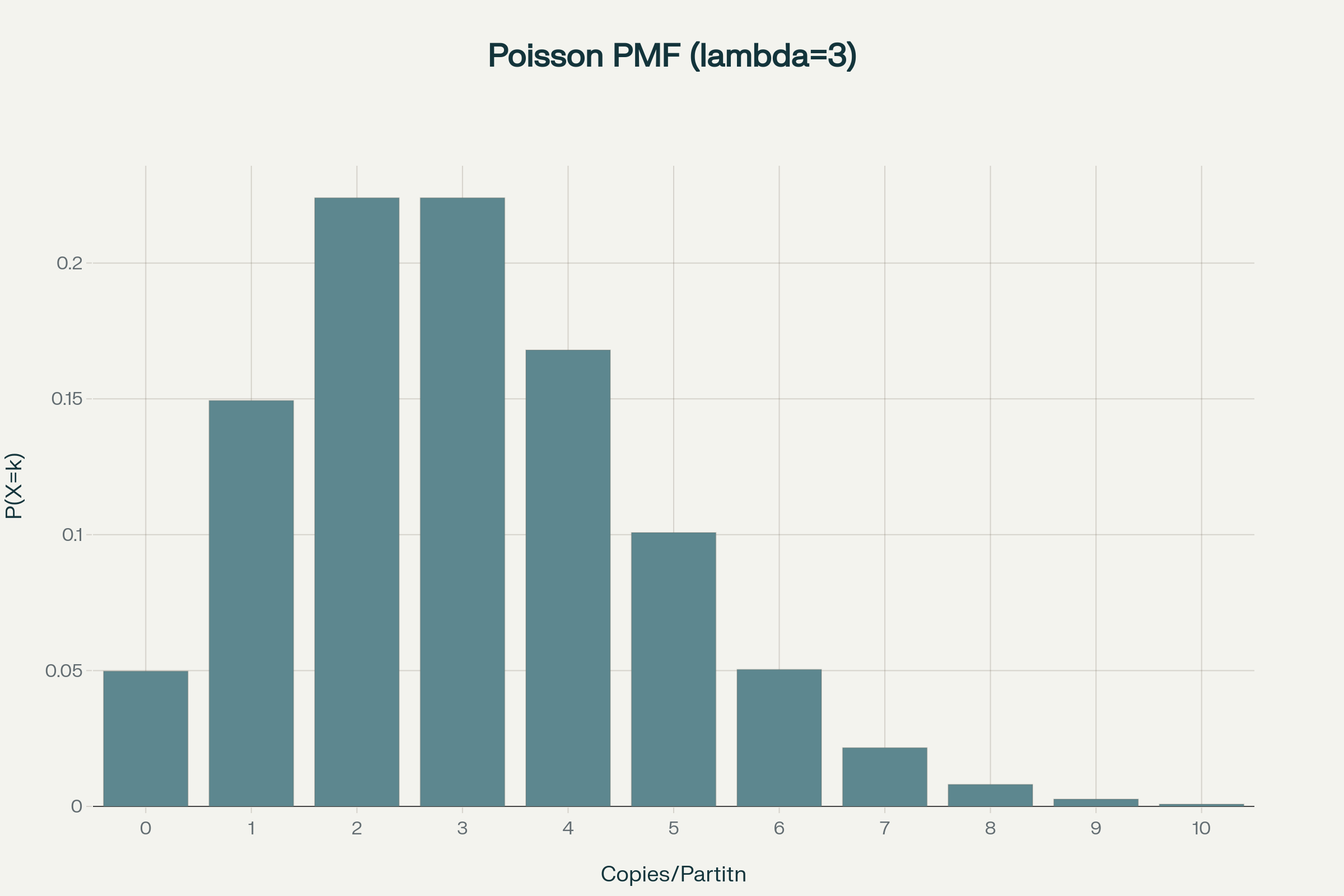
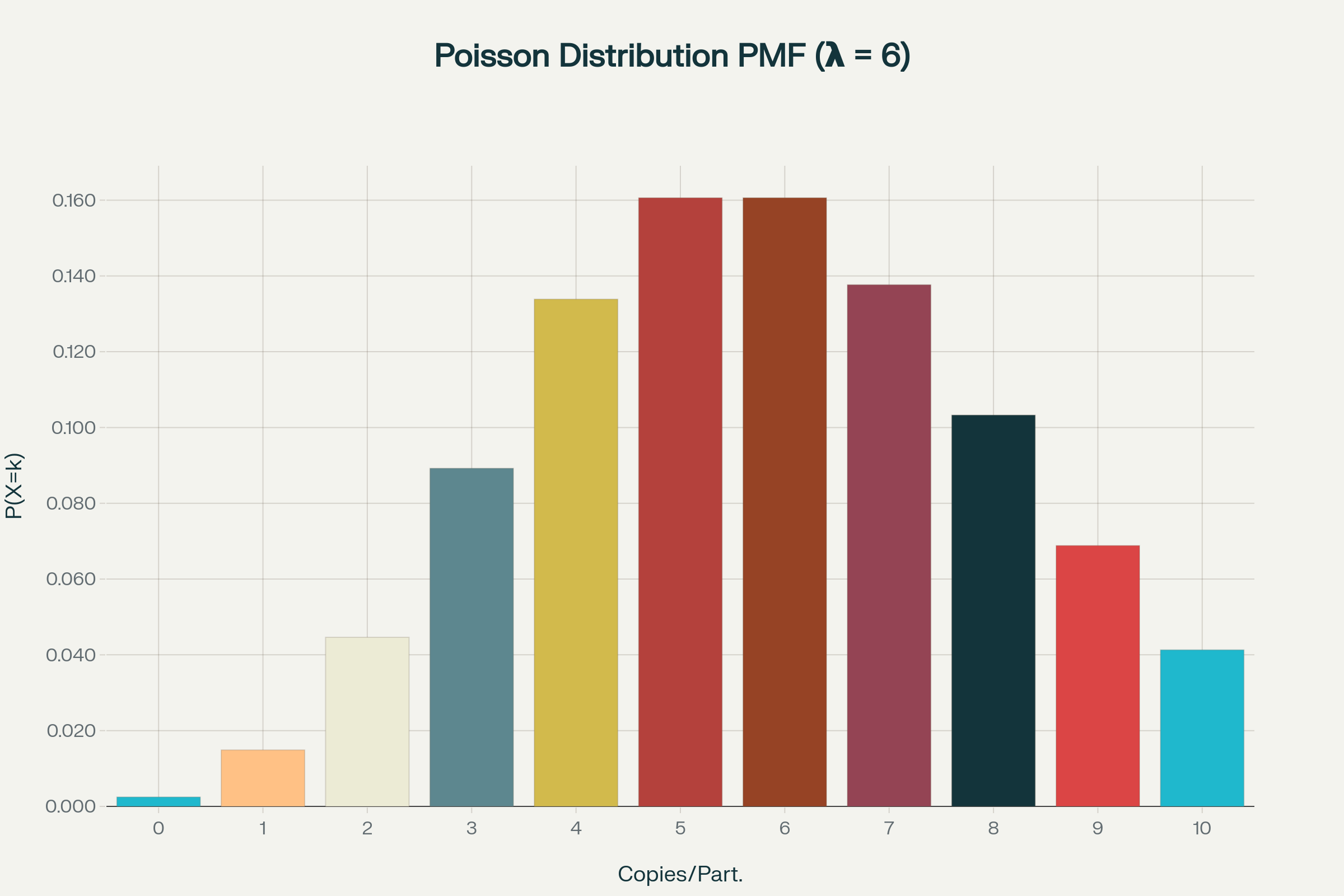
How to calculate copies/droplet and copies/µL in digital PCR?
The total number of copies of the target molecule in all valid droplets of a sample is calculated by multiplying the copies of the target molecule per droplets with the number of valid droplets. Based on the known number of copies of the target molecule per droplet (λ) and the droplet volume, the copies per microliter can also be calculated.

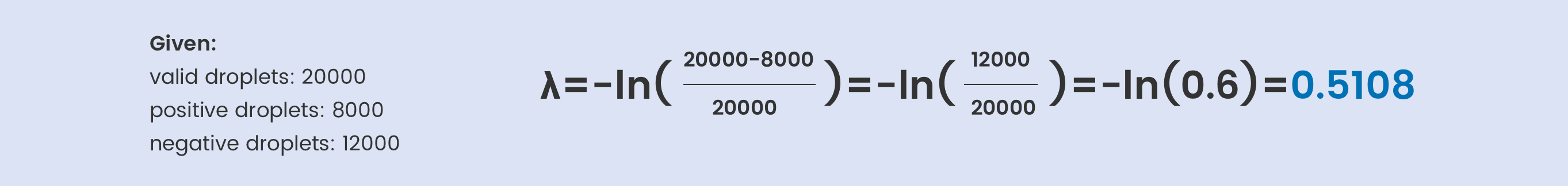


What is the workflow of digital PCR DropDX series?
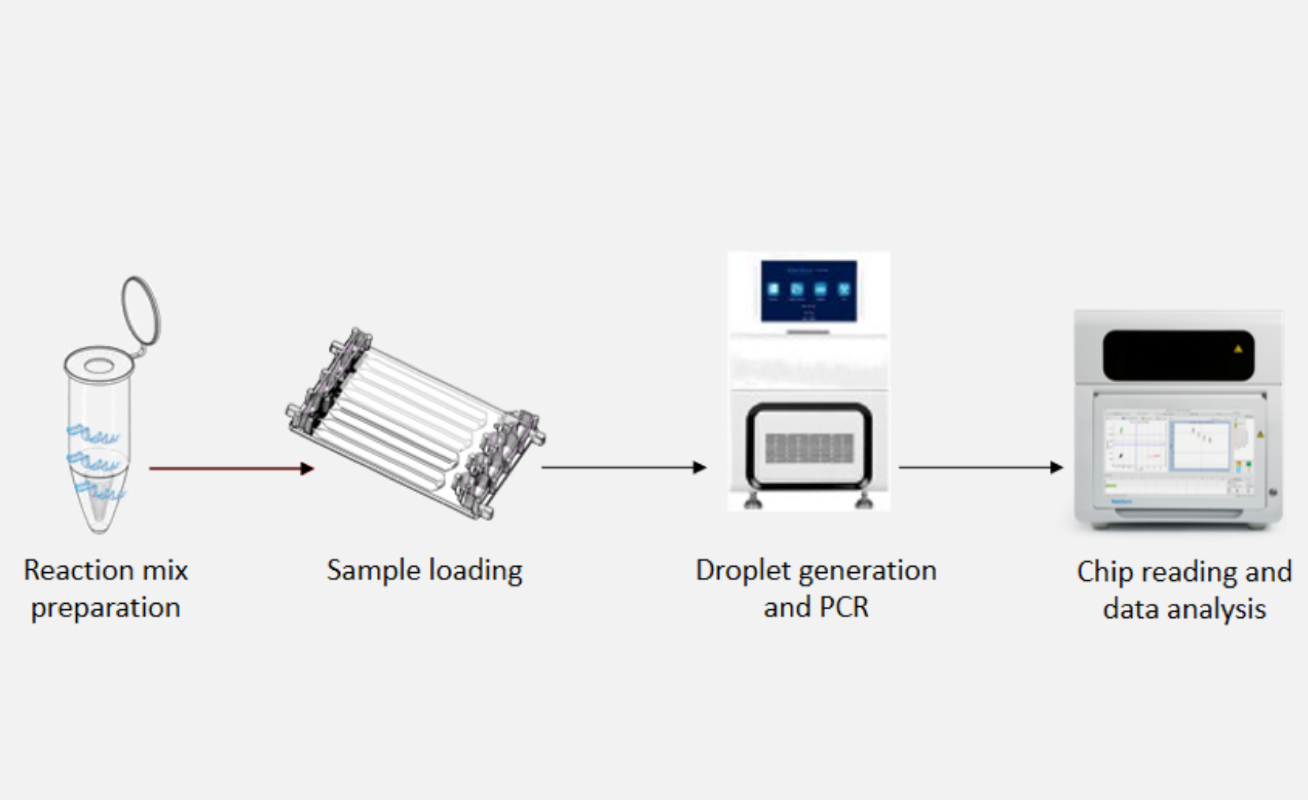
The digital PCR (dPCR) detection workflow begins with nucleic acid extraction from the sample to isolate DNA or RNA. After extraction, the purified nucleic acids are mixed with PCR mastermix, including primers, fluorescent probes, nucleotides, enzymes, and a specialized supermix designed for droplet formation.
What is the workflow of digital PCR RS32 series?
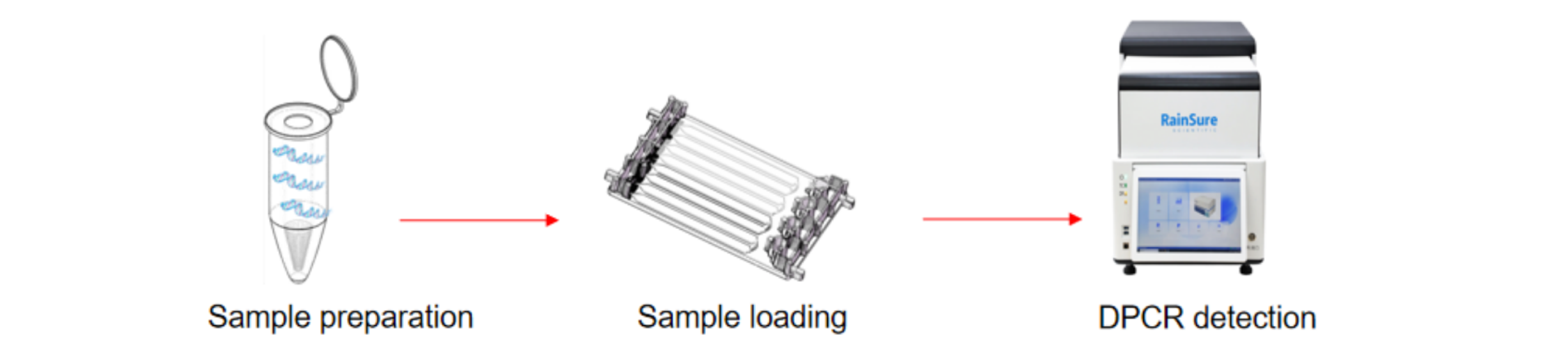
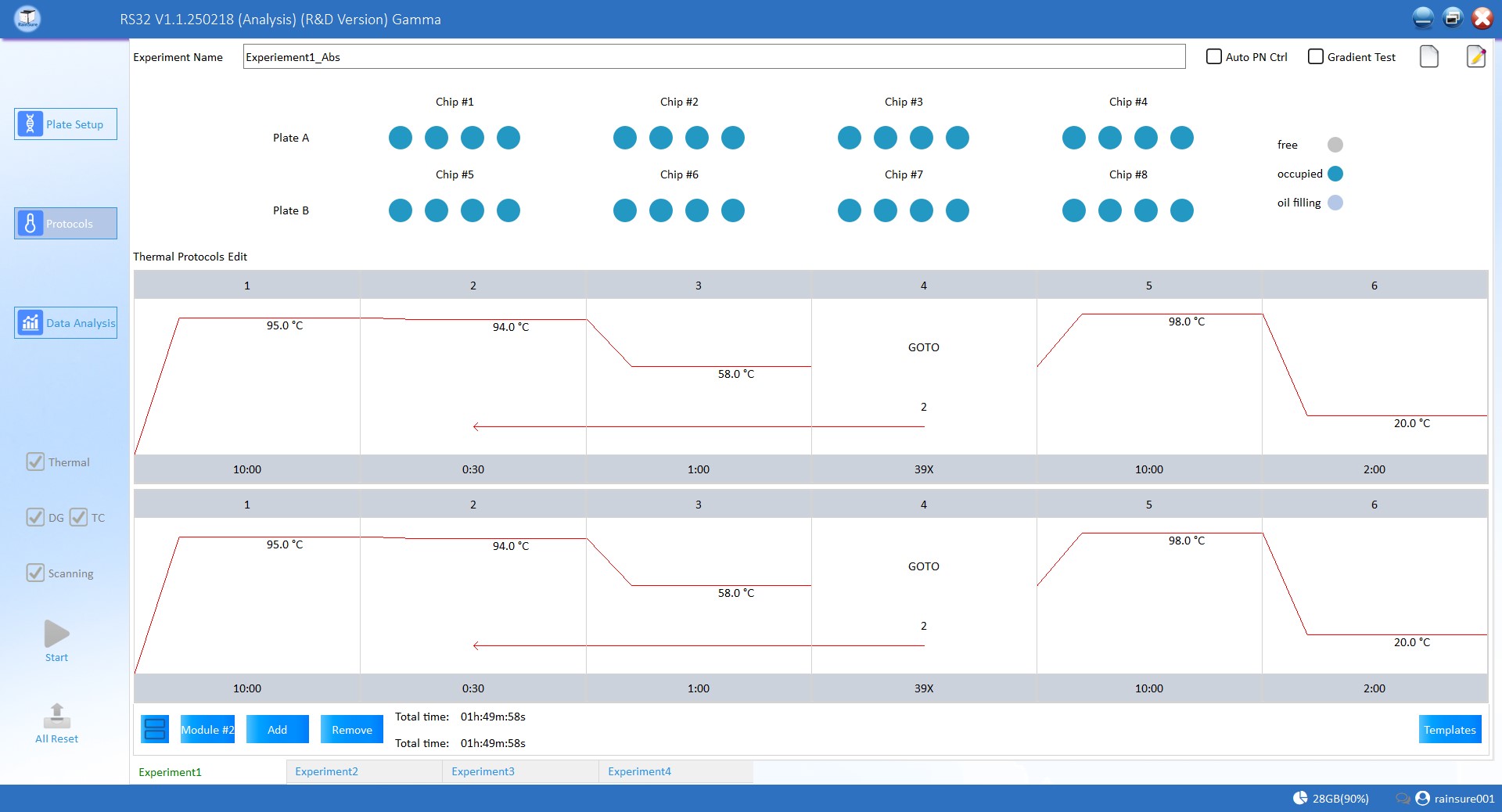
Advantages and limitations of digital PCR
Advantages:
| Advantage | Description |
| Absolute quantification | Directly counts target molecules without the need for a standard curve or reference, improving reliability. |
| High precision and reproducibility | Provides more precise and reproducible results, especially for low-abundance targets and small fold changes. |
| Superior sensitivity | Detects extremely low concentrations of targets, such as rare mutations, trace pathogens, and ctDNA. |
| High tolerance to inhibitors | Partitioning reduces the impact of PCR inhibitors, allowing robust quantification even in challenging samples. |
| No reliance on amplification efficiency | Results are less affected by variations in PCR efficiency, leading to more accurate quantification. |
Disadvantages:
| Disadvantage | Description |
| Narrower dynamic range | The limited number of partitions restricts the range of target concentrations that can be accurately quantified, often requiring sample dilution for highly abundant targets. |
| Higher cost | Equipment and consumables for dPCR are generally more expensive than those for qPCR. |
| Lower throughput | dPCR typically processes fewer samples per run. |
| Limited reagent kit variety | Commercialized reagent kits are still limited in variety, and even fewer kits have obtained qualification for use in hospitals. |
| Non-interchangeable reagent kits | Because the partitioning methods differ, reagent kits from different digital PCR manufacturers are not interchangeable. |
Comparison Between Digital PCR and qPCR
Comparative Table: qPCR vs. dPCR
| Feature/Aspect | Real-time PCR (qPCR) | Digital PCR (dPCR) |
| Quantification | Relative or absolute (requires standard curves or references) | Absolute (no standards or references required) |
| Reaction Format | Bulk PCR, flexible reaction volumes | Sample partitioning, fixed partition volumes |
| PCR Efficiency Impact | Impacted by changes in PCR efficiency (data collected during exponential phase) | Unaffected by amplification efficiency changes |
| Tolerance to Inhibitors | Prone to inhibitors | Higher inhibitor tolerance |
| Detection Method | Real-time detection | End-point detection |
| Dynamic Range | Broad dynamic range | Narrower dynamic range |
| Sensitivity | Higher variation, less sensitive to rare targets | Detects small changes and rare targets, higher sensitivity |
| Reproducibility | Moderate, can be affected by various factors | Higher reproducibility |
| Statistical Power | Lower | Higher |
| Protocol Maturity | Well established protocols | Easy transition from qPCR, but protocols still developing |
| Throughput | High | Lower |
Comparison Between Digital PCR (dPCR) and Next-Generation Sequencing (NGS)
NGS is ideal for broad discovery, identifying a wide range of genetic variants and biomarkers without prior knowledge of mutations. It is fundamental for comprehensive dPCR provides ultrasensitive validation and precise quantification of specific targets, making it suitable for tracking known mutations or rare variants over time, especially in monitoring treatment response.
| NGS | Digital PCR | Digital PCR |
| Detection Limit | 10%-5% for WGS and WES | 0.1%-0.001% |
| Data Analysis | Extensive bioinformatics support, demanding data interpretation | Straightforward |
| Mutation Knowledge Requirement | Prior knowledge of mutations not required | Required |
| Scope of Detection | Hundreds to whole exome or whole genome | Limited |
| Quantification | Semi-quantitative | Absolute quantification |
| Types of Alterations Detected | Copy number alterations, structural rearrangements, SNV or methylation changes over whole genome/panels | Possible on single genes/regions |
| Turnaround Time (TAT) | Long | Short |
| Cost | High | Low |
These technologies are often integrated in workflows: NGS for discovery and profiling, followed by dPCR for sensitive, quantitative monitoring of selected targets. This synergy is particularly valuable in precision medicine, oncology, infectious disease monitoring, and genetic testing.
Applications of Digital PCR
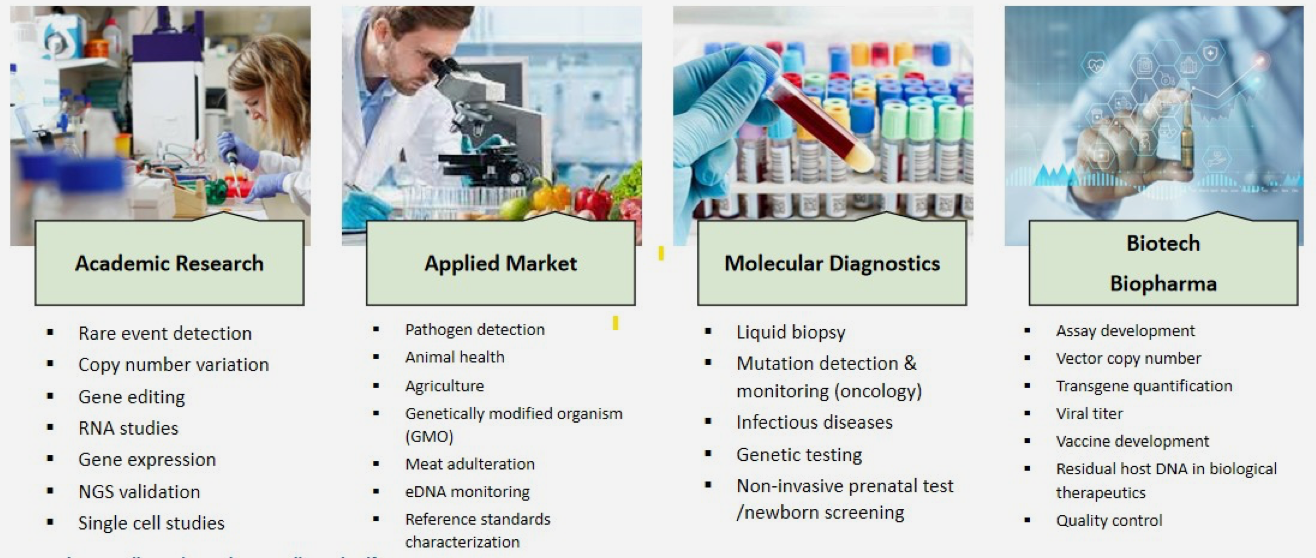
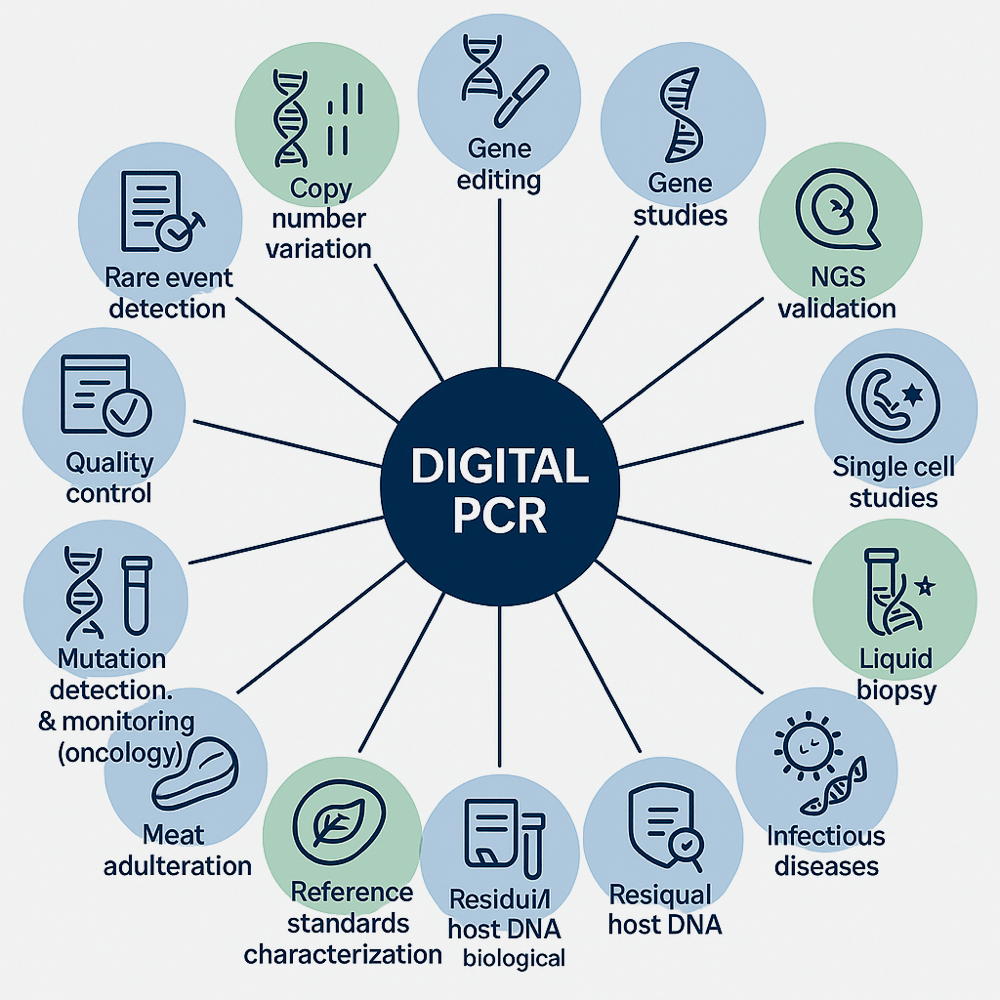
Citations
1.Mu H, Zou J, Zhang H. (2024). Quantitative Detection of T315I Mutations of BCR::ABL1 using Digital Droplet Polymerase Chain Reaction. Hematol Transfus Cell Ther. 17:S2531-1379(24)00030-0. doi: 10.1016/j.htct.2023.12.007.
2.Zhao Z, Wang Y, Kang Y, et al. (2024). A Retrospective Study of the Detection of Sepsis Pathogens Comparing Blood Culture and Culture-independent Digital PCR. Heliyon. 10(6):e27523. doi: 10.1016/j.heliyon.2024.e27523.
3.Mao S, Lin Y, Qin X, et al. (2024). Droplet Digital PCR: An Effective Method for Monitoring and Prognostic Evaluation of Minimal Residual Disease in JMML. Br J Haematol. 204(6):2332-2341. doi: 10.1111/bjh.19465.
4.Chen J, Liu X, Zhang Z, et al. (2024). Early Diagnostic Markers for Esophageal Squamous Cell Carcinoma: Copy Number Alteration Gene Identification and cfDNA Detection. Lab Invest. 104(10):102127. doi: 10.1016/j.labinv.2024.102127.
5.He Y, Dong L, Yan W, et al. (2024). A Multiplex PCR System for the Detection and Quantification of Four Genetically Modified Soybean Events. Resear Square. doi: 10.21203/rs.3.rs-4766822/v1.
6.Dong L, Li W, Xu Q, et al. (2023). A Rapid Multiplex Assay of Human Malaria Parasites by Digital PCR. Clin Chim Acta. 539:70-78. doi: 10.1016/j.cca.2022.12.001.
7.Kopylova KV, Kasparov EW, Marchenko IV, et al. (2023). Digital PCR as a Highly Sensitive Diagnostic Tool: a Review . Mol Bio (Mosk). 57(5):771-781. doi: 10.31857/S0026898423050051. [Article in Russian]
8.Lu R, Wang J, Li M, et al. (2022). Retrospective Quantitative Detection of SARS-CoV-2 by Digital PCR Showing High Accuracy for Low Viral Load Specimens. J Infect Dev Ctries. 16(1), 10-15. doi: 10.3855/jidc.15315.
Related kit
| CAT No. | Name |
| 3.02.03.0001 | BCR ABL p210/p190 digital PCR one-tube quant test |
| 3.02.03.0005 | PIK3CA 11 Mutations Digital PCR Assay |
| 3.02.03.0006 | Human JAK2 Gene V617F Mutation Detection Kit (Digital PCR Method) |
| 3.02.03.0008 | Malaria 18S rRNA gene detection kit (digital PCR) |
| 3.02.03.0009 | Fastplex™ BCR-ABL (p210) %IS digital PCR Kit |
| 3.02.01.4066 | Digital PCR Mastermix for Probes |
| 3.02.01.4071 | Human ALK Gene G1269A Mutation Detection Assay |
| 3.02.01.4072 | Human BRAF Gene V600E Mutation Detection Assay |
| 3.02.01.4073 | Human EGFR Gene G465R Mutation Detection Assay |
| 3.02.01.4074 | Human EGFR Gene G719S Mutation Detection Assay |
| 3.02.01.4075 | Human EGFR Gene G719A Mutation Detection Assay |
| 3.02.01.4076 | Human EGFR Gene E746_A750delELREA (COSM6223) Mutation Detection Assay |
| 3.02.01.4077 | Human EGFR Gene E746_A750delELREA (COSM6225) Mutation Detection Assay |
| 3.02.01.4078 | Human EGFR Gene L747_A750delinsP (COSM2238) Mutation Detection Assay |
| 3.02.01.4079 | Human EGFR Gene L747_A750delinsP (COSM2239) Mutation Detection Assay |
| 3.02.01.4080 | Human EGFR Gene L747_P753>S Mutation Detection Assay |
| 3.02.01.4081 | Human EGFR Gene S768I Mutation Detection Assay |
| 3.02.01.4082 | Human EGFR Gene V769_D770insASV Mutation Detection Assay |
| 3.02.01.4083 | Human EGFR Gene D770_N771insG Mutation Detection Assay |
| 3.02.01.4084 | Human EGFR Gene T790M Mutation Detection Assay |
| 3.02.01.4085 | Human EGFR Gene C797S (COSM6493937) Mutation Detection Assay |
| 3.02.01.4086 | Human EGFR Gene C797S (COSM5945664) Mutation Detection Assay |
| 3.02.01.4087 | Human EGFR Gene L858R Mutation Detection Assay |
| 3.02.01.4088 | Human EGFR Gene L861Q Mutation Detection Assay |
| 3.02.01.4089 | Human ESR1 Gene Y537S Mutation Detection Assay |
| 3.02.01.4090 | Human ESR1 Gene D538G Mutation Detection Assay |
| 3.02.01.4091 | Human KRAS Gene G12D Mutation Detection Assay |
| 3.02.01.4092 | Human KRAS Gene G12V Mutation Detection Assay |
| CAT No. | Name |
| 3.02.01.4093 | Human KRAS Gene G12C Mutation Detection Assay |
| 3.02.01.4094 | Human KRAS Gene G12A Mutation Detection Assay |
| 3.02.01.4095 | Human KRAS Gene G12S Mutation Detection Assay |
| 3.02.01.4096 | Human KRAS Gene G13D Mutation Detection Assay |
| 3.02.01.4097 | Human NRAS Gene G12V Mutation Detection Assay |
| 3.02.01.4098 | Human NRAS Gene Q61R Mutation Detection Assay |
| 3.02.01.4099 | Human PIK3CA Gene E542K Mutation Detection Assay |
| 3.02.01.4100 | Human PIK3CA Gene E545K Mutation Detection Assay |
| 3.02.01.4101 | Human PIK3CA Gene H1047R Mutation Detection Assay |
| 3.02.01.4102 | Human TP53 Gene R175H Mutation Detection Assay |
| 3.02.01.4103 | Human TP53 Gene R248L Mutation Detection Assay |
| 3.02.01.4104 | Human TP53 Gene R273C Mutation Detection Assay |
| 3.02.01.4105 | Human TP53 Gene R273H Mutation Detection Assay |
| 3.02.01.4106 | Human HER2 Gene Copy Number Variation Detection Assay |
| 3.02.01.4108 | Human influenza A virus H1N1 -2009 Detection Assay |
| 3.02.01.4109 | Human influenza A virus H3N2 Detection Assay |
| 3.02.01.4113 | Human Epstein-Barr Virus (BamHI-W) Detection Assay |
| 3.02.01.4114 | Human Epstein-Barr Virus (EBNA1) Detection Assay |
| 3.02.01.4115 | Human CMV Detection Assay |
| 3.02.01.4117 | Human BCR-ABL T315I Mutation Detection Assay |
| 3.02.01.4120 | Human IDH1 Gene R132C Mutation Detection Assay |
| 3.02.01.4121 | Human IDH1 Gene R132G Mutation Detection Assay |
| 3.02.01.4122 | Human IDH1 Gene R132H Mutation Detection Assay |
| 3.02.01.4123 | Human MYD88 Gene L265P Mutation Detection Assay |
| 3.02.01.4130 | Human BCR-ABL (P210) Fusion Gene Detection Kit (Digital PCR) |
| 3.02.01.4138 | Human BCR-ABL P190 Detection Assay |
| 3.02.01.4265 | SARS-CoV-2 Detection Kit( RT-dPCR) |
| 3.02.01.4277 | Sepsis pathogenic microorganism detection kit(digital PCR) |







